Authors

Independent Health, like most healthcare insurers, continually navigates a labyrinth of compliance requirements and documentation. One crucial document for policyholders is the "Certificate of Coverage" (CoC), which details the extent, limitations, and conditions under which a healthcare provider will offer coverage. It's a complex document outlining benefits, coverage limits, and the procedures for filing claims. Understanding and managing a CoC can be daunting both for insurers and customers due to its complexity, often spanning over a hundred pages with intricate sections, sub-sections, and tables.
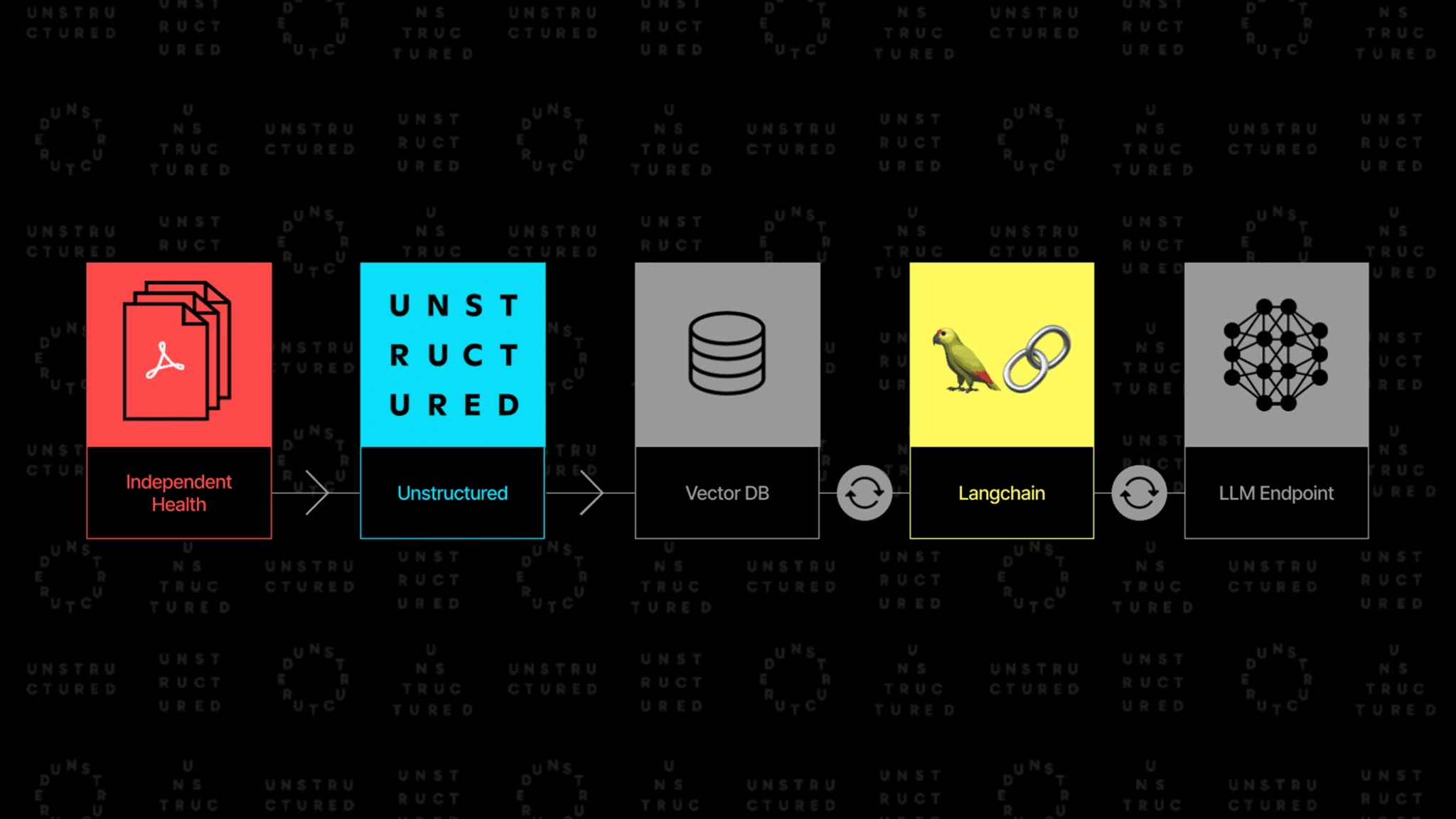
Recognizing the need for managing these extensive documents with clarity and efficiency, LangChain and Unstructured collaborated with Independent Health of New York to explore how a Retrieval-Augmented Generation (RAG)-based architecture could be leveraged to make it easier to quickly answer questions about insurance policies.
The insights outlined below underscore the value of leveraging Unstructured and LangChain to optimize RAG architectures to process semi-structured content from Independent Health’s CoC. In the context of a RAG architecture, semi-structured data combines elements of both structured and unstructured data, typically involving structured tables interspersed with natural language text, such as in PDF file formats, which require specialized preprocessing.
We compare a text-only Baseline RAG to a nuanced Semi-Structured RAG architecture enhanced by Unstructured and LangChain tools. We leverage Unstructured’s data ingestion and preprocessing capabilities to transform semi-structured data into LLM-friendly formats. We also use LangChain’s Multi-Vector Retrieval to orchestrate data retrieval. Using LangChain’s evaluation platform, LangSmith, we assess the performance of both architectures. To do so, we prompt each with the same set of four questions, comparing their responses to the expected solution. Baseline RAG succeeded in only one of the four cases, while Semi-Structured RAG successfully answered three of the four questions.
Disclaimer: The data in this blog post has been altered for confidentiality. We have used replacement data, subjected to the same rigorous analysis as actual data, to ensure the integrity and relevance of our insights.
Innovative Tools for Transforming Semi-Structured Data
Unstructured brings cutting-edge preprocessing and transformation tools. These are designed to ingest and process the most challenging technical documents and convert them into a structured JSON format that’s ML-ready.
Complementing this, LangChain's Multi-Vector Retriever tool is a game-changer; it decouples documents intended for answer synthesis from references used for retrieval. A prime example of its utility is summarizing verbose documents to optimize them for vector-based similarity search. It simultaneously ensures that the entire document is passed into the Large Language Model (LLM), preserving every nuance and preventing any loss of context during answer synthesis. This two-pronged approach ensures that healthcare providers can navigate the CoC quickly, ultimately enhancing the service they provide to policyholders.
Tackling the Challenges of Semi-Structured Data in RAG Architecture
Characterized by a combination of text and tables, semi-structured data doesn't conform to the rigidity of structured databases nor the free-form nature of unstructured data. Traditional RAG systems often stumble when faced with semi-structured documents for two primary reasons:
1. Text Splitting: They often inadvertently disrupt the integrity of tables during text splitting, leading to incomplete data retrieval.
2. Table Embedding: RAG context generally depends on semantic similarity search to query relevant content in a database. This proves particularly challenging in the case of tables because tables are not just plain text; they are structured data where the significance of each piece of information is often determined by its position (row and column) and its relationship to other data points in the table.
We address these challenges with a combination of Unstructured’s ingestion and preprocessing transformation tools and LangChain’s Multi-Vector Retriever to enhance the capabilities of a traditional RAG architecture when handling semi-structured data.
Data Ingestion and Processing from PDFs
Accurate parsing of documents is the first step to address the challenges of semi-structured data. The Independent Health documents for this case study are in PDFs. These documents largely depend on elements like a table of contents, lists, and tables; elements known to cause trouble for conventional PDF partitioning packages. See examples below.
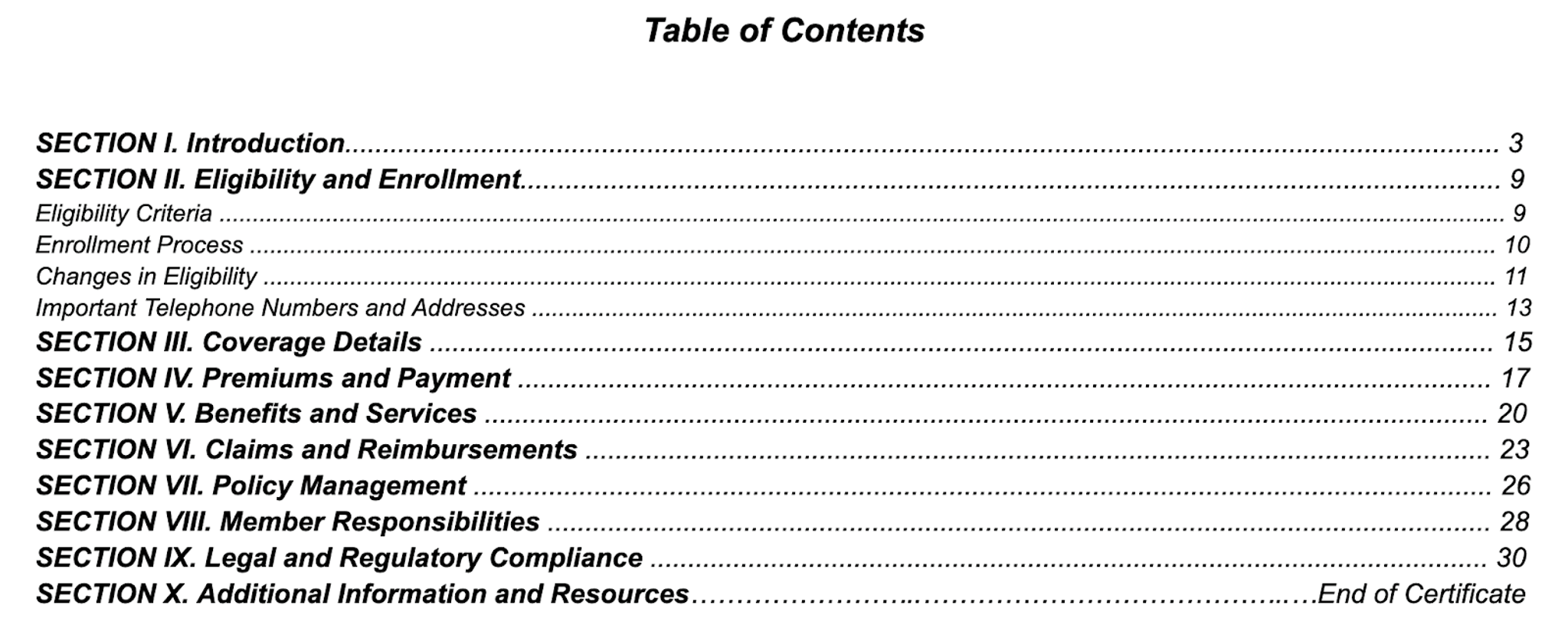
1. Table of Contents
The image below shows the Table of Contents from a health insurance policy manual. It includes section titles describing different aspects of policy coverage, each linked to a right-aligned page number through dot leaders for visual alignment. Here, OCR struggles to handle variable image quality, interpret formatting and fonts correctly, and differentiate between titles and page numbers.
- Nested List Items
Ingesting data from nested list structures involves several challenges:
- OCR technology must accurately recognize and differentiate between the hierarchy of main list items and any sub-items.
- Variations in document formatting hinder consistent recognition of list items.
- Downstream semantic retrieval requires an association between inline list items and their surrounding context.

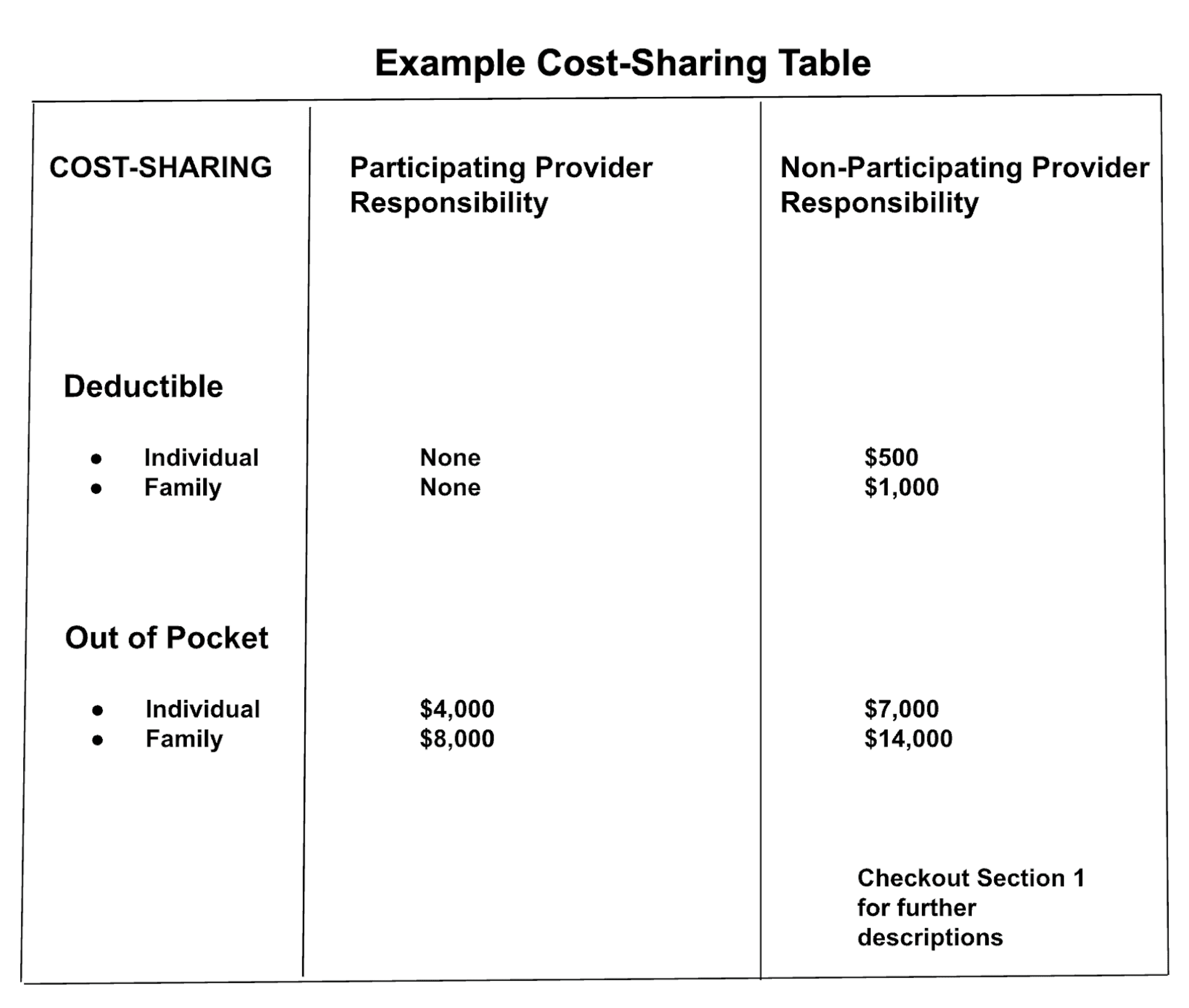
3. Complex Tables
In the simplest case, a table adheres to a uniform grid of rows in columns. This is generally straightforward to extract. However, in practice, tables are often more complex than that. An example is provided below. Ingesting data from these complex tables involves detecting and grouping values by section. In some cases, ancillary notes or instructions are placed outside the standard grid layout. These must be associated with the corresponding data points within the table. OCR systems must also navigate varying text formats, such as bold headings and asterisks indicating footnotes or additional information. This can affect the integrity of the data extraction process.
Unstructured preprocessing tools are fine-tuned to address these kinds of concerns explicitly. With partition_pdf, we transform a raw PDF document into JSON, making the tables and text machine-readable. It also attributes rich metadata to the text and tables, which is particularly helpful in post-processing and downstream NLP tasks. The code below does just that.
The output is a list of Elements which each contain the corresponding document text and all associated metadata. Unstructured identifies and tags all Elements by their type (e.g., text vs table…etc). This allows users or downstream processes to filter and operate based on this property. The following code shows how to extract specific elements by type.
Storing Documents: A Tale of Two Approaches
With the data extracted, the focus shifts to storage; note, this significantly impacts retrieval performance. We explore two distinct approaches:
Approach 1: Multi-Vector Retriever
Using a multi-vector retriever to store raw tables and text alongside the summaries, which is more conducive to retrieval, helps to address the complexity of semi-structured data. This strategically structures the storage to facilitate more effective retrieval.
Approach 2: Baseline Vector Store (No Table Retrieval)
The Baseline Vector Store represents the traditional approach. Documents are stored without any explicit consideration of tables. This method acts as the control in our evaluation, providing a benchmark against which we compare to the multi-vector retriever.
We now can query either approach by using the respective chain’s invoke function.
Building the Evaluation Dataset
LangSmith is a platform for building production LLM applications. It also has embedded capabilities to test and evaluate them. To assess the performance of our approaches, we first construct an evaluation set in LangSmith using question-and-answer (QA) pairs. An example of this and its output can be seen below.
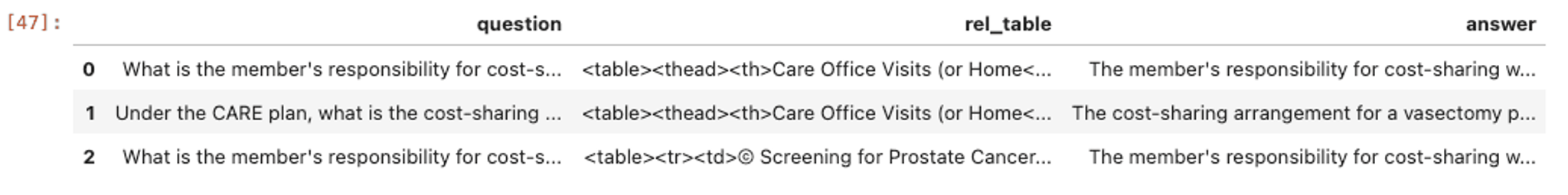
Dataframe of the Evaluation Dataset
After constructing the QA pair, we upload the dataset to LangSmith.
The Evaluation Process
The evaluation process employs a systematic approach to measure and compare the performance of the Baseline and Semi-Structured RAGs. This process is crucial in quantifying the benefits of the multi-vector retriever and table partitioning strategy.
The complete LangSmith evaluation workflow is shown in the figure below.
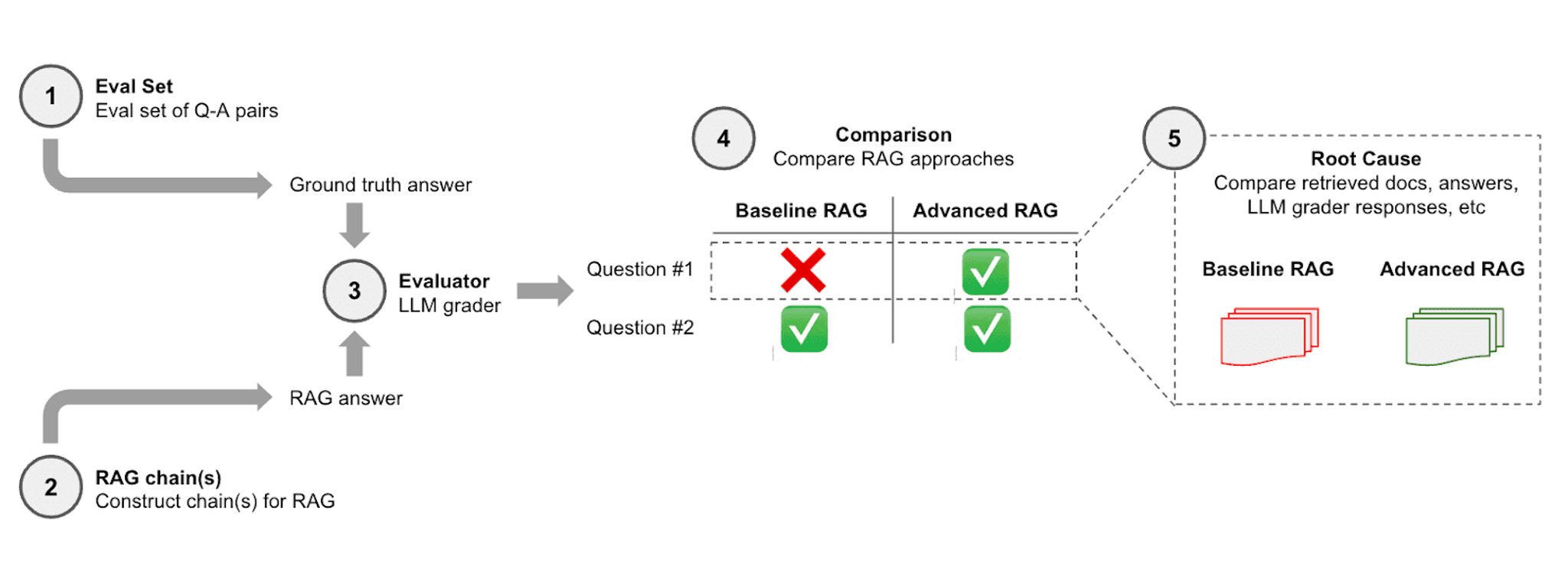
LangSmith compares baseline and advanced models through a series of steps:
- Eval Set (Evaluation Set): This step involves the collection of question-and-answer (Q-A) pairs that will be used to test the RAG systems. The answers are considered the ground truth against which the RAG systems' outputs will be compared.
- RAG chain(s): The RAG constructs chains of reasoning or steps to arrive at an answer for each question from the evaluation set. This represents the RAG systems' attempt to generate answers.
- Evaluator (LLM grader): A Large Language Model (LLM) grader assesses the answers generated by the RAG systems by comparing them to the ground truth answers from step 1.
- Comparison: The performance of the Baseline RAG and Advanced RAG is compared based on their answers. For example, the Baseline RAG failed to answer Question #1 correctly but succeeded with Question #2. The Advanced RAG answered both questions correctly.
- Root Cause: This final step involves a deeper analysis of why each RAG model succeeded or failed. It involves comparing retrieved documents, the answers, and LLM grader responses.
LangSmith offers various evaluators for RAG that can determine the relevance of a model's response for common evaluation scenarios. In this case, we use the QA evaluators to measure the correctness of a response to a user query or question. Specifically, the "qa" evaluator (reference) instructs an LLMChain to directly grade a response as "correct" or "incorrect" based on the reference answer.
LangSmith’s `run_on_dataset` function will run the Chain or Language Model on a dataset and store traces to the specified project name. Here is how to run the process:
For further information about LangSmith, check out this cookbook as a reference.
Comparative Analysis: A Clear Winner Emerges
In the test, we prompted both Baseline RAG and Semi-Structured RAG with the same questions. We evaluated their responses against each of the expected correct solutions. Of the four questions, Baseline RAG only successfully answered one of these. Semi-Structured RAG provided correct responses to three of the four questions, a clear improvement. A deeper investigation into the nature of the responses shows the efficacy of the multi-vector retriever approach and the importance of tailoring storage and retrieval strategies to accommodate the nature of semi-structured data.
Analysis
In this section, we step through some of the questions asked and responses given by both the Baseline and Semi-Structured RAGs. We refer back to the source documents for an explanation as to how each arrived at their answers.
Question 1: Semi-Structured Answered Correctly
Question
What is the member's responsibility for cost-sharing when using a non-participating provider for OBGYN preventative care?
Baseline RAG Answer
The document does not provide specific information on the member's responsibility for cost-sharing when using a non-participating provider for OBGYN preventative care?
Semi-Structured RAG Answer
The member's responsibility for cost-sharing when using a non-participating provider for OBGYN preventative care is 40% Coinsurance after Deductible.
Reference Table
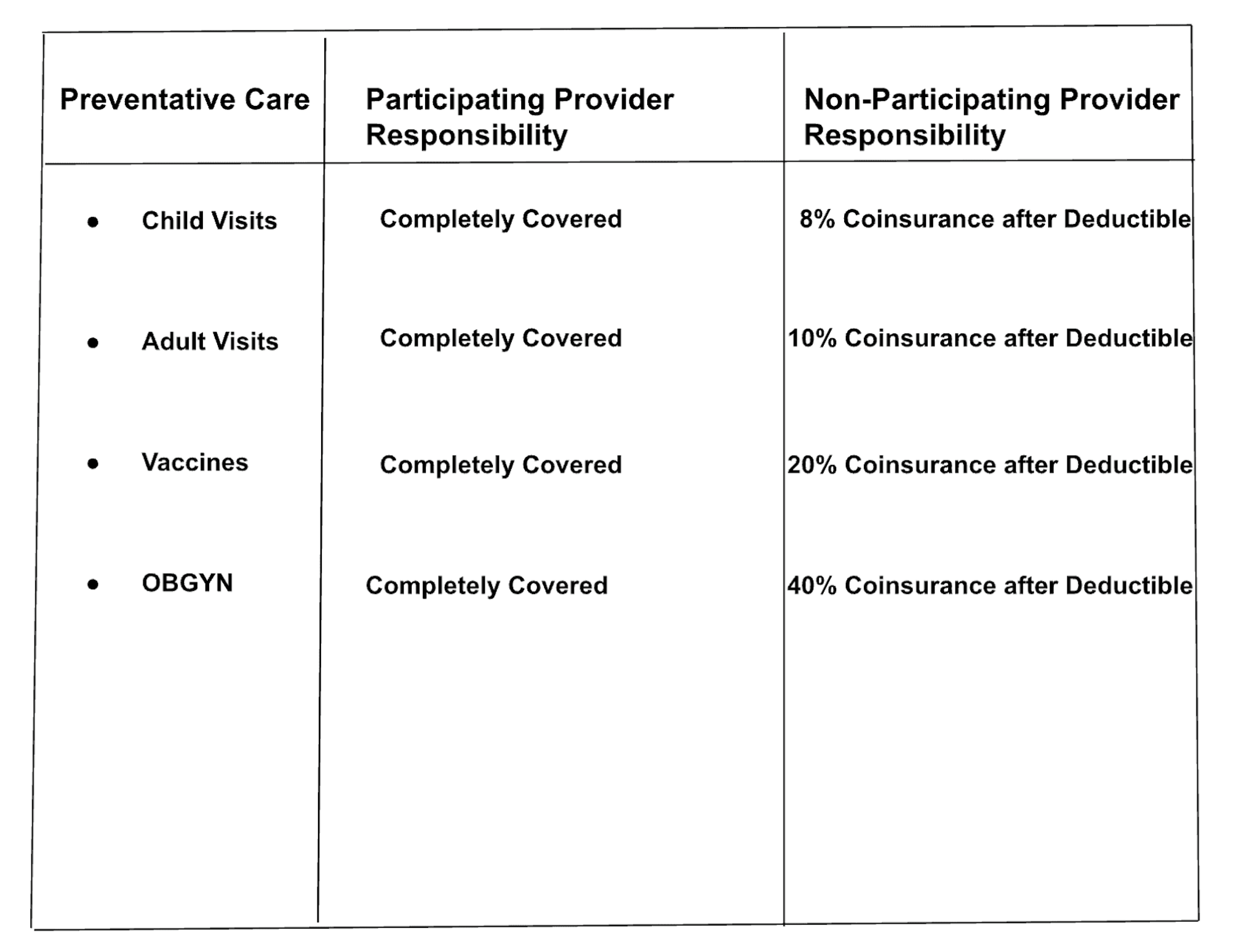
Analysis
Baseline RAG does not retrieve any critical, relevant table information. As a result, it responds with a misleading representation that the document is missing this information. Made possible by Unstructured preprocessing, Semi-Structured RAG retrieves the entire table as a single record in the vector database. This provides RAG with the complete context necessary to answer the question correctly.
Question 2: Both Approaches Answered Incorrectly
Question
What is the cost-sharing arrangement for an ENT procedure when using a participating provider?
Baseline RAG Answer
The document does not provide specific information on the cost-sharing arrangement for an ENT procedure when using a participating provider.
Semi-Structured RAG Answer
The cost-sharing arrangement for an ENT procedure when using a participating provider is covered in full.
Correct Answer
The cost-sharing arrangement for an ENT procedure when using a participating provider is to "Use Cost-Sharing for appropriate service (Deviated Septum;Hearing Test;Aesthesia)."
Reference Table
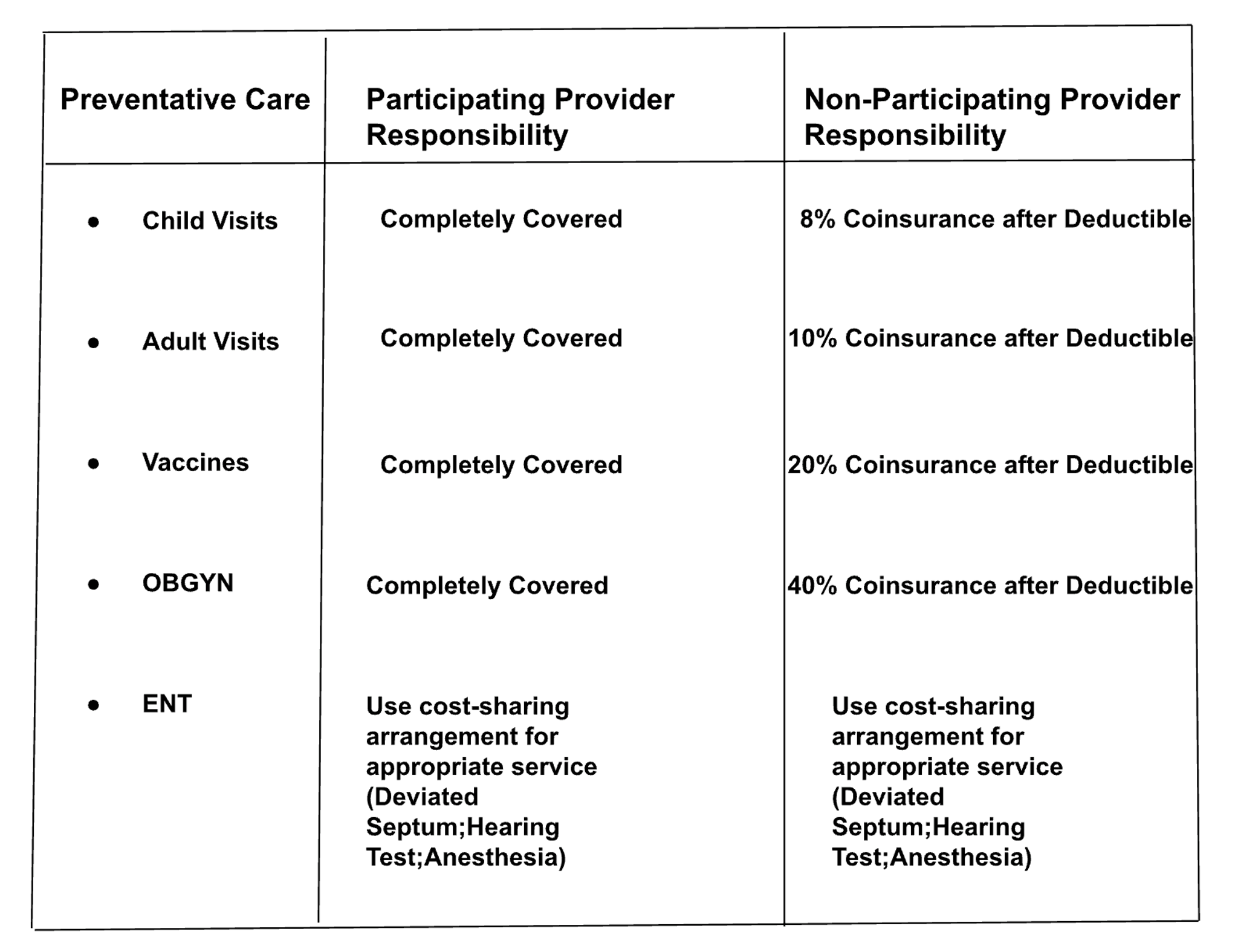
Analysis
Baseline RAG and Semi-Structured RAG are incorrect but for different reasons. The Baseline RAG retriever doesn’t pull any information from the relevant table — including text elements; in fact, it references text from otherwise unrelated pages. Semi-structured RAG correctly identifies the table. However, the table structure is not as clean and, as a result, answers the question incorrectly. This highlights the importance of clean data extraction and room for even more improvements Unstructured should make to its underlying models to reconstruct table format and cells.
Question 3: Both Approaches Answered Correctly
Question
What is the member's responsibility for cost-sharing when preventive services are not provided in accordance with the HIPAA?
Baseline RAG Answer
The member's responsibility for cost-sharing when preventive services are not provided in accordance with HIPAA is to use cost-sharing for the appropriate service. This could include a PCP Visit, Specialist Visit, Blood Work, Sleep Test.
Semi-Structured RAG Answer
The member's responsibility for cost-sharing when preventive services are not provided in accordance with HIPAA is to use cost-sharing for the appropriate service. This could include a PCP Visit, Specialist Visit, Blood Work, Sleep Test.
Reference Table
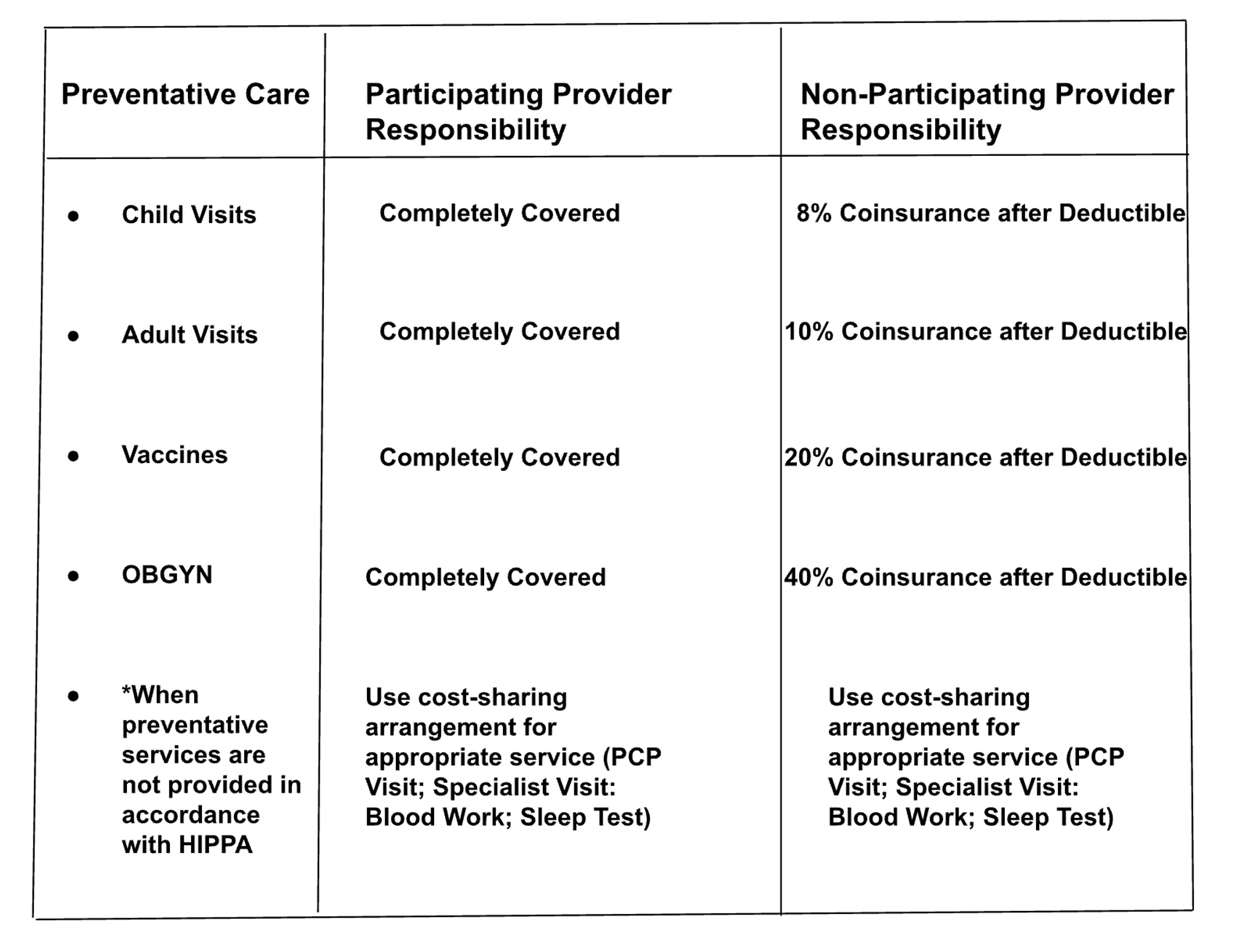
Analysis
Both approaches answer this question correctly by retrieving the correct table. This question is notably more specific and is only addressed in the reference table, which is why Baseline RAG can also find the correct document. In the Semi-Structured case, the correct table is the first result returned, implying higher similarity.
Conclusion
The comparative analysis of Baseline and Semi-Structured RAG approaches reveals key insights into their respective capabilities in handling semi-structured data. The examples showcase the Semi-Structured RAG's proficiency in accurately utilizing table partitioning compared to the Baseline RAG. This underlines the transformative impact of semi-structured data handling techniques in RAG architecture.
The challenges of semi-structured data in RAG architecture are non-trivial but surmountable. Tools like Unstructured for document parsing and the LangChain Multi-Vector Retriever for intelligent data storage are key to our approach's success. Collectively, these tools enable enhanced retrieval accuracy beyond traditional RAG applications. Such advancements set a precedent for future data retrieval and processing endeavors.
About Unstructured
Unstructured is a leading provider of LLM data preprocessing solutions, empowering organizations to transform their internal unstructured data into formats compatible with large language models. By automating the transformation of complex natural language data found in formats like PDFs, PPTX, HTML files, and more, Unstructured enables enterprises to leverage the full power of their data for increased productivity and innovation. With key partnerships and a growing customer base of over 10,000 organizations, Unstructured is driving the adoption of enterprise LLMs worldwide. Questions? Connect with us directly in our community Slack.
About LangChain
LangChain is a framework designed for building applications that leverage language models to be context-aware and capable of sophisticated reasoning. It offers a suite of tools, including LangChain Packages, LangChain Templates, LangServe, and LangSmith. Check out LangChain resources, including the Quickstart guide and Discord community.
About Independent Health
Independent Health is an independent, not-for-profit health plan headquartered in Buffalo, New York, providing innovative health care products and benefits designed to engage consumers in their health and well-being. Established in 9180, Independent Health offers a comprehensive portfolio of progressive products including HMO, POS, PPO and EPO products, Medicare Advantage and Medicaid plans, individual Exchange products, consumer-directed plans, health savings accounts, and coverage for self-funded employers. Our subsidiaries and affiliate companies include a third-party administrator of health benefits, pharmacy benefit management, specialty pharmacy and the Independent Health Foundation. In all, Independent Health and its affiliates serve a total of more than 550,000 lives across the country. Independent Health has been recognized nationally for its award-winning customer service, dedication to quality health care and unmatched relationships with physicians and providers. To learn more, go to www.independenthealth.com.